For more than half a century, many biologists have leaned on the neutral theory of molecular evolution to explain how DNA and proteins change over time. The idea grew from early work in the 1960s, when scientists began sequencing proteins and later genes, shifting attention from outward traits to the genetic code itself. The model suggested that most mutations that become fixed in a population are neither helpful nor harmful. Harmful ones are quickly filtered out and helpful ones are so rare that, in theory, nearly all long-term genetic change looks neutral.
A new study from the University of Michigan paints a more complex picture. It shows that helpful mutations are far more common than expected, yet very few of them take over a population. You see an outcome that looks neutral even though the underlying process is anything but.
The work, published in Nature Ecology and Evolution and supported by the National Institutes of Health, introduces a new way to think about how living things respond to a world that never stops shifting.
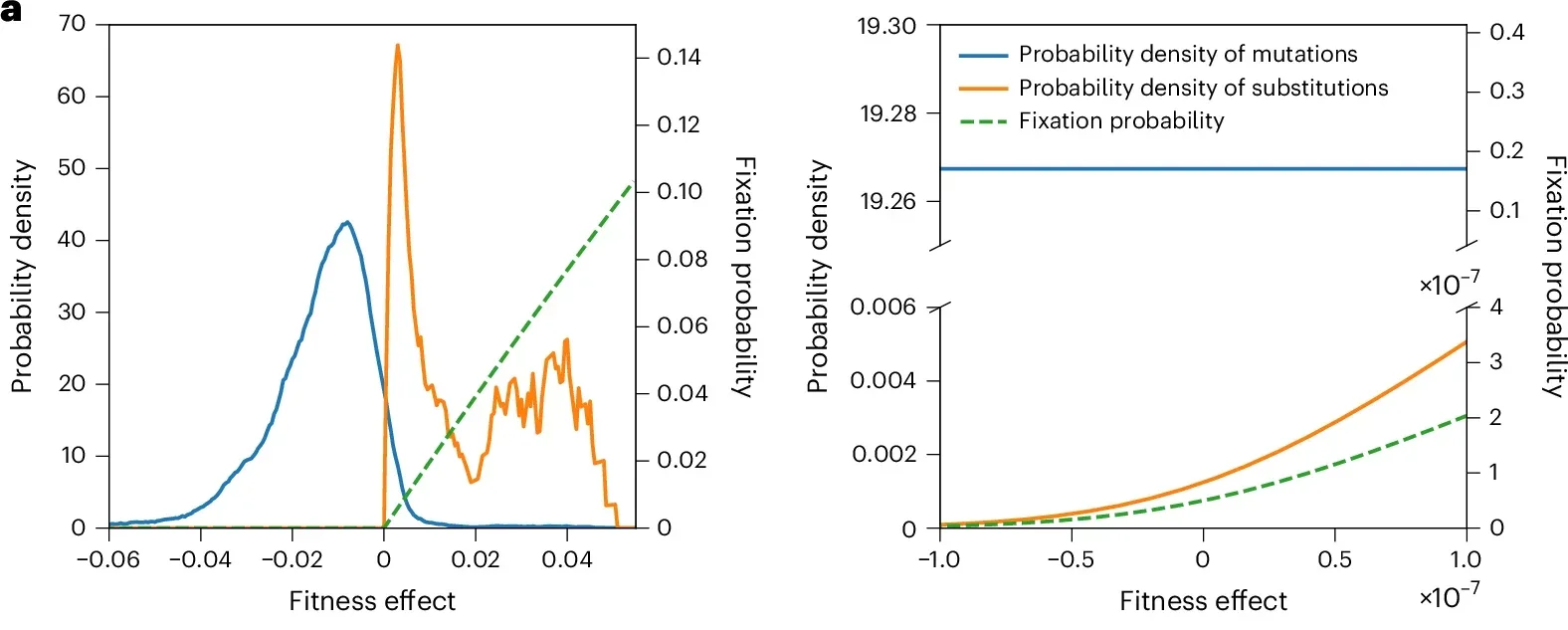
The research team, led by evolutionary biologist Jianzhi Zhang, used deep mutational scanning to measure how new amino acid changes influence the fitness of yeast and bacteria. This method lets scientists alter a gene in thousands of ways and test how each change affects growth over many generations. By comparing mutated strains with normal ones, the team could estimate which mutations were harmful, neutral or helpful.
Across genes from yeast and E. coli, more than one percent of all tested amino acid changes improved fitness. That may seem small at first, but it is orders of magnitude higher than what the neutral theory allows. If those mutations occurred in stable environments, more than 99 percent of all long-term genetic changes should be adaptive. In reality, long-term data from nature do not show such rapid adaptive evolution. Something was missing.
The team realized that most earlier models assumed the environment stays the same for long stretches of time. But real conditions do not offer that kind of stability. Temperature, nutrients, predators and countless other pressures shift in ways that populations cannot fully predict. A mutation that helps in one situation can hurt in the next.
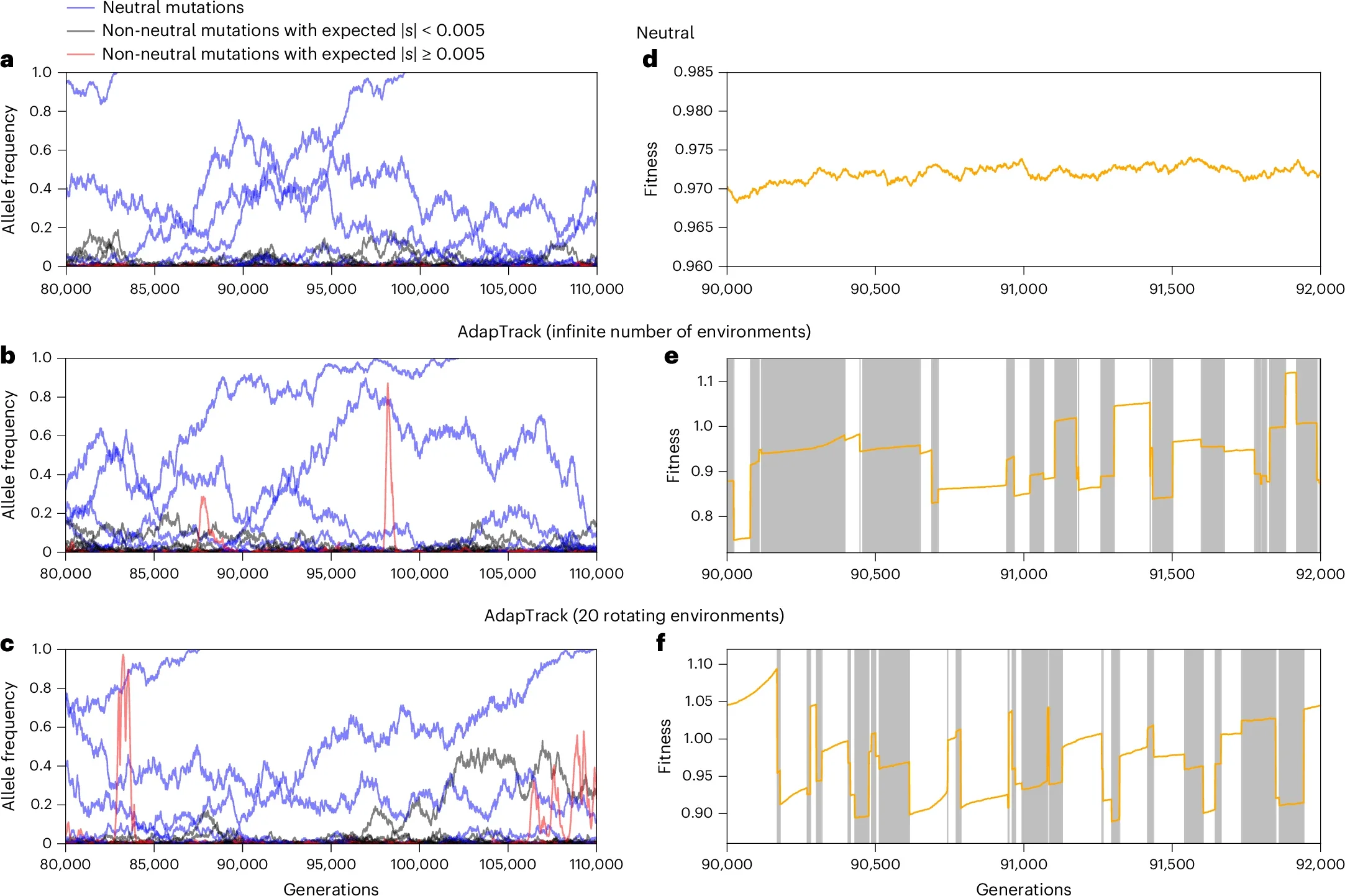
Zhang’s group tested this idea with a massive yeast experiment. One set of populations grew in the same environment for 800 generations. Another set cycled through ten different environments, each for 80 generations, for the same total time. The difference was striking. In the stable setting, beneficial mutations piled up. In the changing setting, far fewer did, even though helpful mutations still appeared. They simply did not have time to spread before the next environmental change flipped their effects.
“This is where the inconsistency comes from,” Zhang said. “While we observe a lot of beneficial mutations in a given environment, those beneficial mutations do not have a chance to be fixed because as their frequency increases to a certain level, the environment changes.”
The team calls this idea “adaptive tracking with antagonistic pleiotropy.” Antagonistic pleiotropy means that a single mutation can have opposite effects across different conditions. In practice, this means a mutation that increases fitness one day may reduce it the next.
Under adaptive tracking, populations are always trying to catch up to their surroundings. Some helpful mutations climb in abundance for a while, but few survive long enough to spread through the entire population. Instead, many are purged when the environment flips again. Mutations that are nearly neutral in most conditions, even if slightly harmful, are more likely to drift to fixation because they rarely become strongly detrimental.
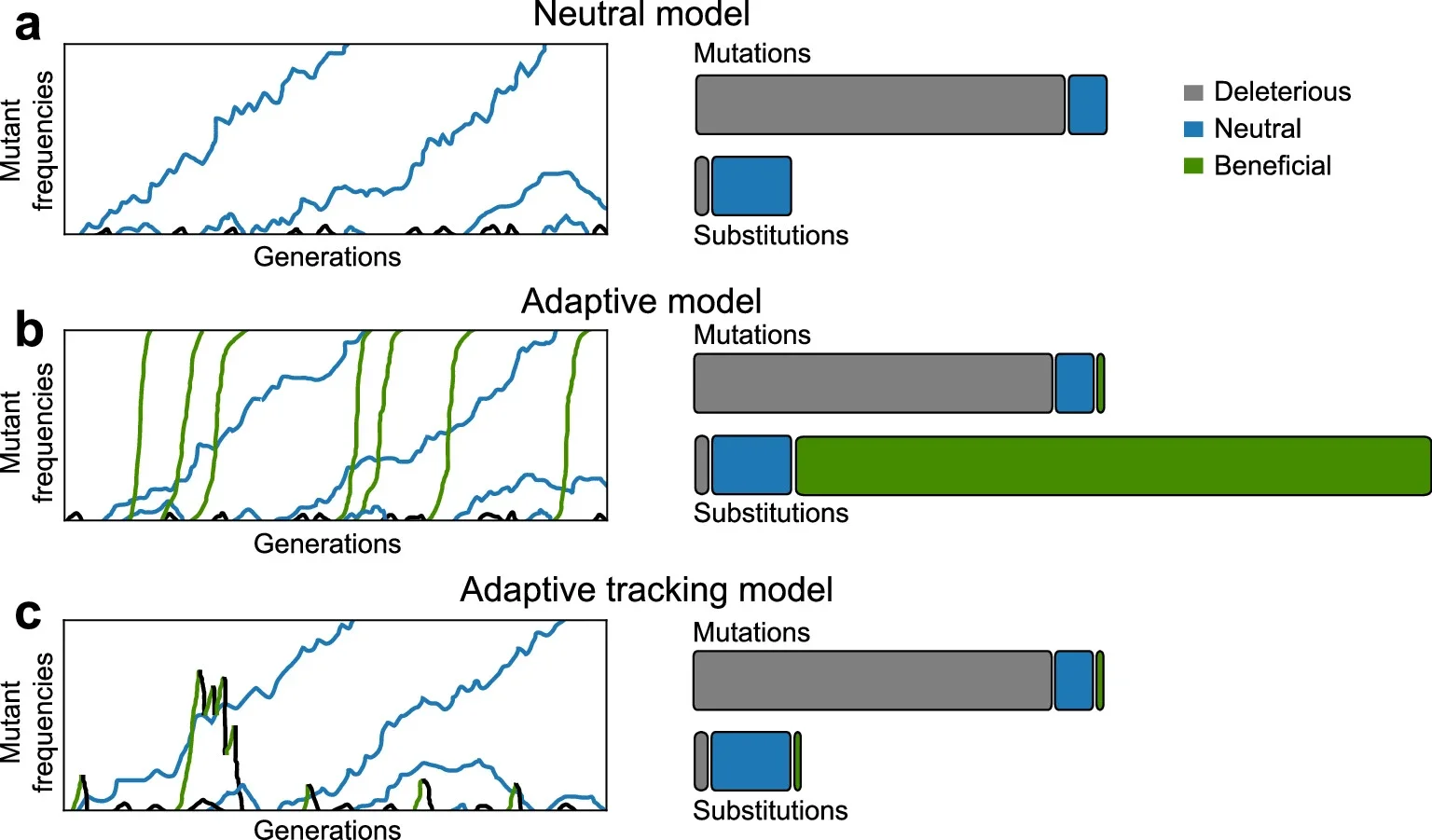
“We’re saying that the outcome was neutral, but the process was not neutral,” Zhang said. “Our model suggests that natural populations are not truly adapted to their environments because environments change very quickly, and populations are always chasing the environment.”
This constant chase helps explain why functional genes often evolve at rates similar to nonfunctional DNA. Even though selection works hard on short time scales, the long-term pattern can look quiet and steady, almost like a molecular clock.
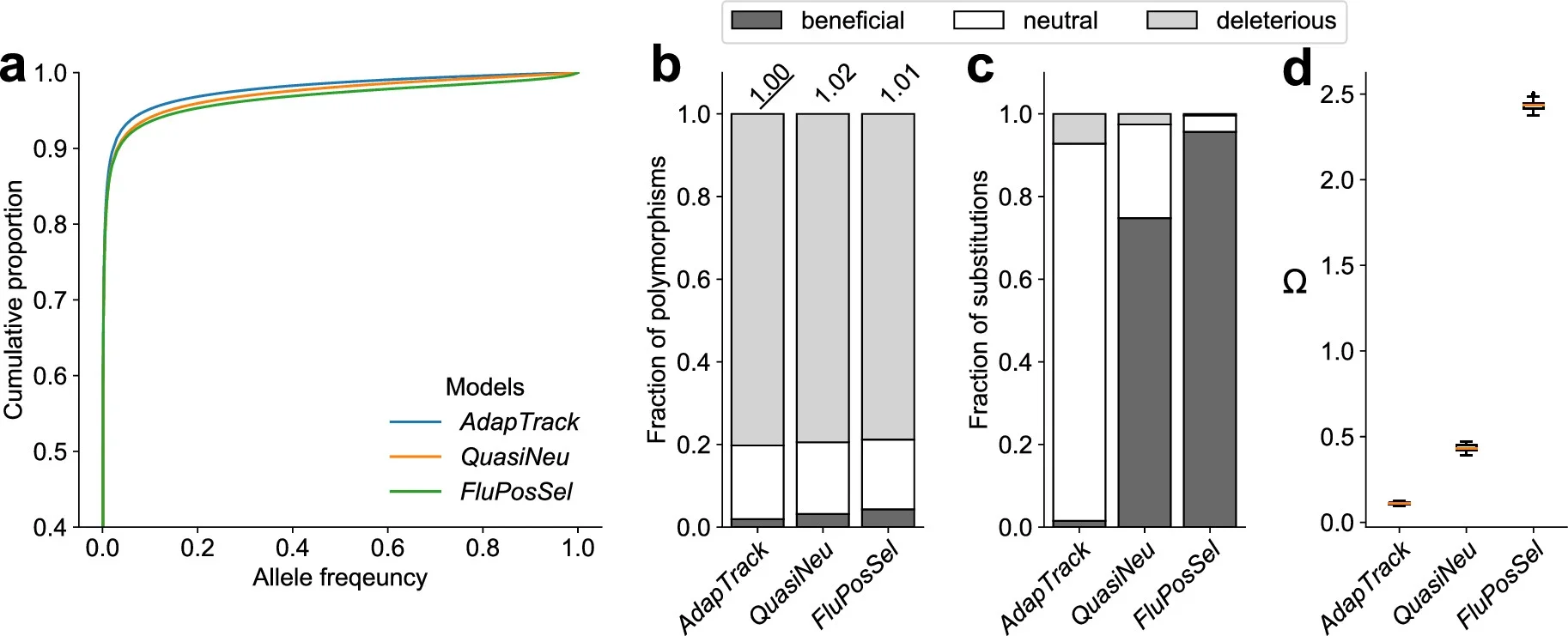
To test how well adaptive tracking explains long-term evolutionary patterns, the team ran large computer simulations. They compared four scenarios: pseudogenes, a classic neutral model, constant-environment adaptation and adaptive tracking with shifting conditions.
Under stable adaptation, helpful mutations quickly dominated, and long-term change was strongly adaptive. Under adaptive tracking and neutral models, however, most long-term substitutions behaved as if they were neutral. Even when many helpful mutations were present, their chance of fixation stayed low in the changing environments.
The simulations also showed that substitution rates remained steady over time, matching the molecular clock pattern that puzzled researchers for decades. This happened even though adaptation was happening on short time scales.
Zhang believes the work sheds light on how living things relate to the environments that shaped them.
“I think this has broad implications,” he said. He noted that human genes reflect many environments that no longer exist. Mutations that once helped may now be mismatched to present-day life.
You see a world where no species ever becomes perfectly adapted because true stability is rare. Populations may appear well matched to their surroundings at one moment and poorly suited the next, depending on when the last major environmental change occurred. Even thousands of generations are often not enough time to fully adapt before conditions shift again.
The team’s next step is to test whether this pattern holds in larger, multicellular organisms, where deep mutational scanning is harder to perform.
Adaptive tracking suggests that long-term genetic patterns can hide the real amount of ongoing selection. This insight may influence how researchers interpret evolutionary histories and predict how species respond to climate change.
It can also help people understand why humans carry genetic traits shaped by past environments that no longer match modern life.
As scientists apply these ideas to more species, the model may improve forecasts of disease evolution, agricultural resilience and conservation planning.
Research findings are available online in the journal Nature Ecology & Evolution.
Like these kind of feel good stories? Get The Brighter Side of News’ newsletter.
The post Researchers reveal a groundbreaking new theory of evolution appeared first on The Brighter Side of News.