Accurately determining the binding sites in protein-protein interactions is essential for breakthroughs in biomedicine. However, existing experimental methods are slow and expensive.
Computational approaches, particularly those integrating artificial intelligence (AI), promise faster solutions but often fall short when predicting interactions involving synthetic proteins.
A recent study, led by Dr. Rafael Bernardi at Auburn University and published in The Journal of the American Chemical Society, addresses these challenges through a novel combination of AI and molecular dynamics simulations, offering a promising pathway for personalized cancer therapies.
Targeting the PD-L1 protein is a cornerstone of many cancer immunotherapies. PD-L1 allows cancer cells to evade immune detection by suppressing immune responses. Drugs blocking PD-L1, such as pembrolizumab, unleash the immune system to attack tumors. However, accurately predicting binding sites for such treatments remains a formidable challenge.
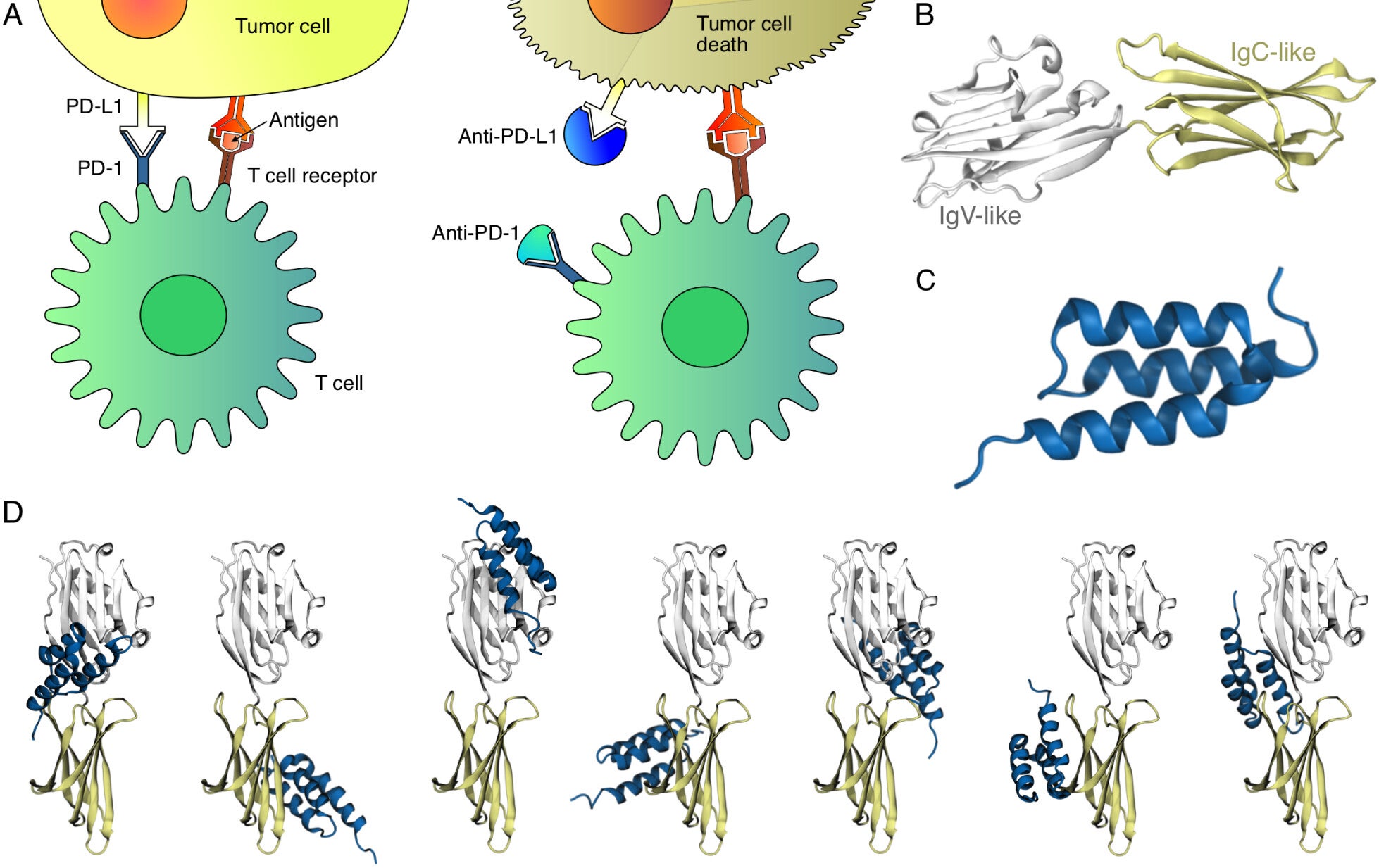
Dr. Bernardi’s team tackled this issue by enhancing AI tools with molecular dynamics simulations and dynamic network analysis. Traditional AI-based methods, including AlphaFold2, were effective but insufficient in distinguishing between binding models.
The team’s integrated approach proved crucial, demonstrating that the perpendicular binding pose of PD-L1:Affibody complexes was significantly more stable than the parallel configuration. This prediction was validated through advanced experimental techniques, including cross-linking mass spectrometry and next-generation sequencing-based mutational scanning.
The study involved researchers from Auburn University, the University of Basel, and ETH Zurich. According to Dr. Bernardi, “Utilizing computational tools to engineer proteins represents the next frontier in cancer therapeutics.” This interdisciplinary collaboration was pivotal in refining computational predictions with experimental validation.
Dr. Diego Gomes, a lead researcher on the project, emphasized the importance of this collaboration: “This work showcases the synergy between computational innovations and experimental techniques, driving breakthroughs in cancer therapy.”
Related Stories
By leveraging cutting-edge resources like NVIDIA DGX systems, the team advanced the capabilities of tools such as NAMD and VMD, underscoring the role of high-performance computing in modern biophysics.
Dynamic network analysis proved to be the cornerstone of this research. By applying this technique to molecular dynamics trajectories generated from AlphaFold2Multimer structures, researchers identified the most stable binding configurations. Notably, AlphaFold3 failed to predict the perpendicular binding pose, highlighting the limitations of AI-only models and the need for integrated methods.
These findings challenge the assumption that AI-generated protein structures can be accepted without rigorous scrutiny. “Our research underscores the necessity of combining AI tools with dynamic network analysis to enhance prediction accuracy,” Dr. Bernardi stated.
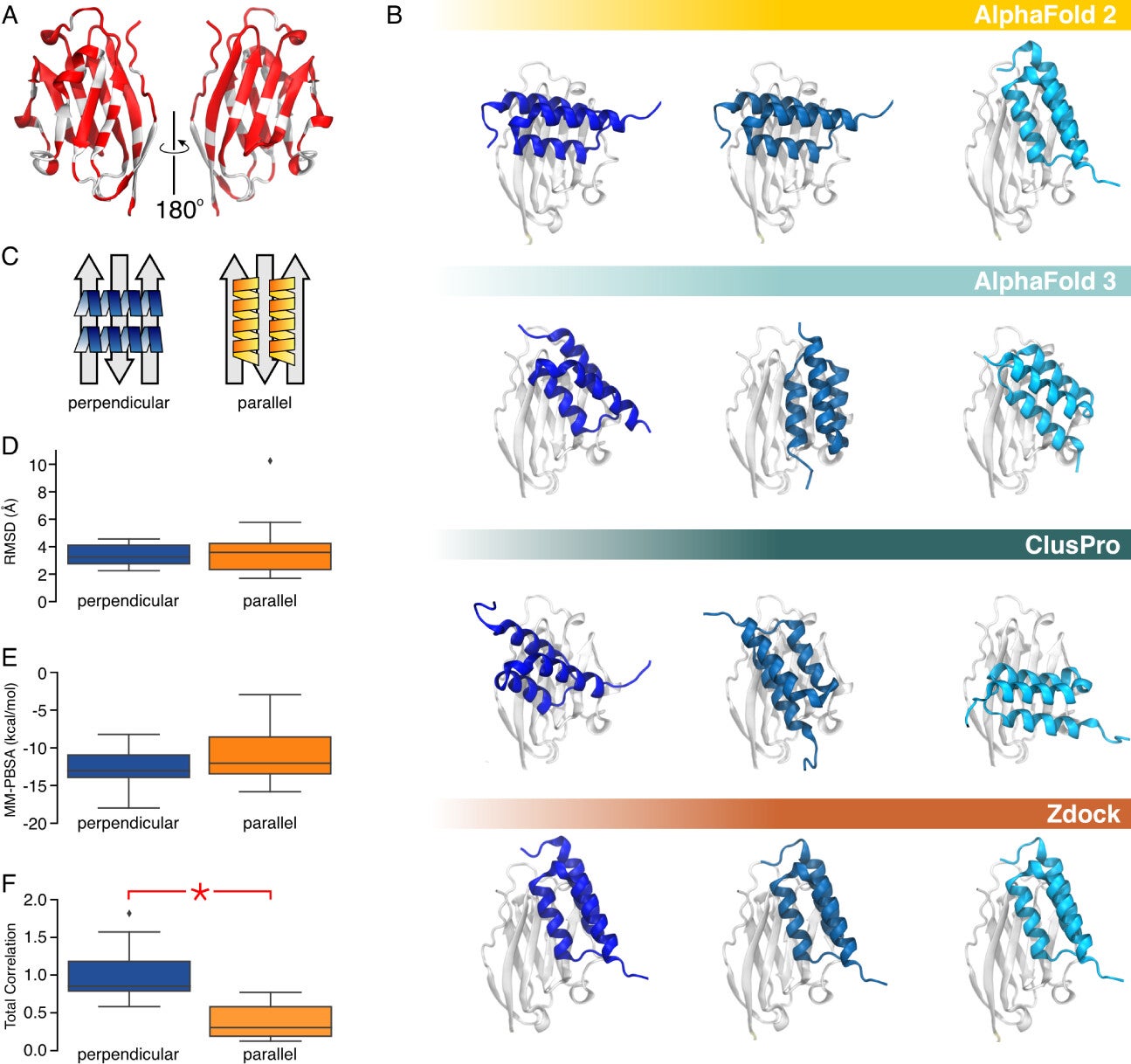
While this study focused on PD-L1, its implications extend far beyond cancer immunotherapy. The methodologies developed can be applied to other proteins, paving the way for new treatments for various diseases, including autoimmune conditions. Additionally, this approach could accelerate drug discovery and reduce costs, addressing limitations of traditional experimental methods.
Dr. Gomes highlighted this broader impact: “The potential applications of our methods are vast, from identifying drug targets in diverse diseases to revolutionizing personalized medicine.”
This work demonstrates how interdisciplinary research can lead to significant advancements in tackling complex medical challenges. By bridging the gap between computational predictions and experimental validation, the study sets a new standard for developing targeted cancer therapies.
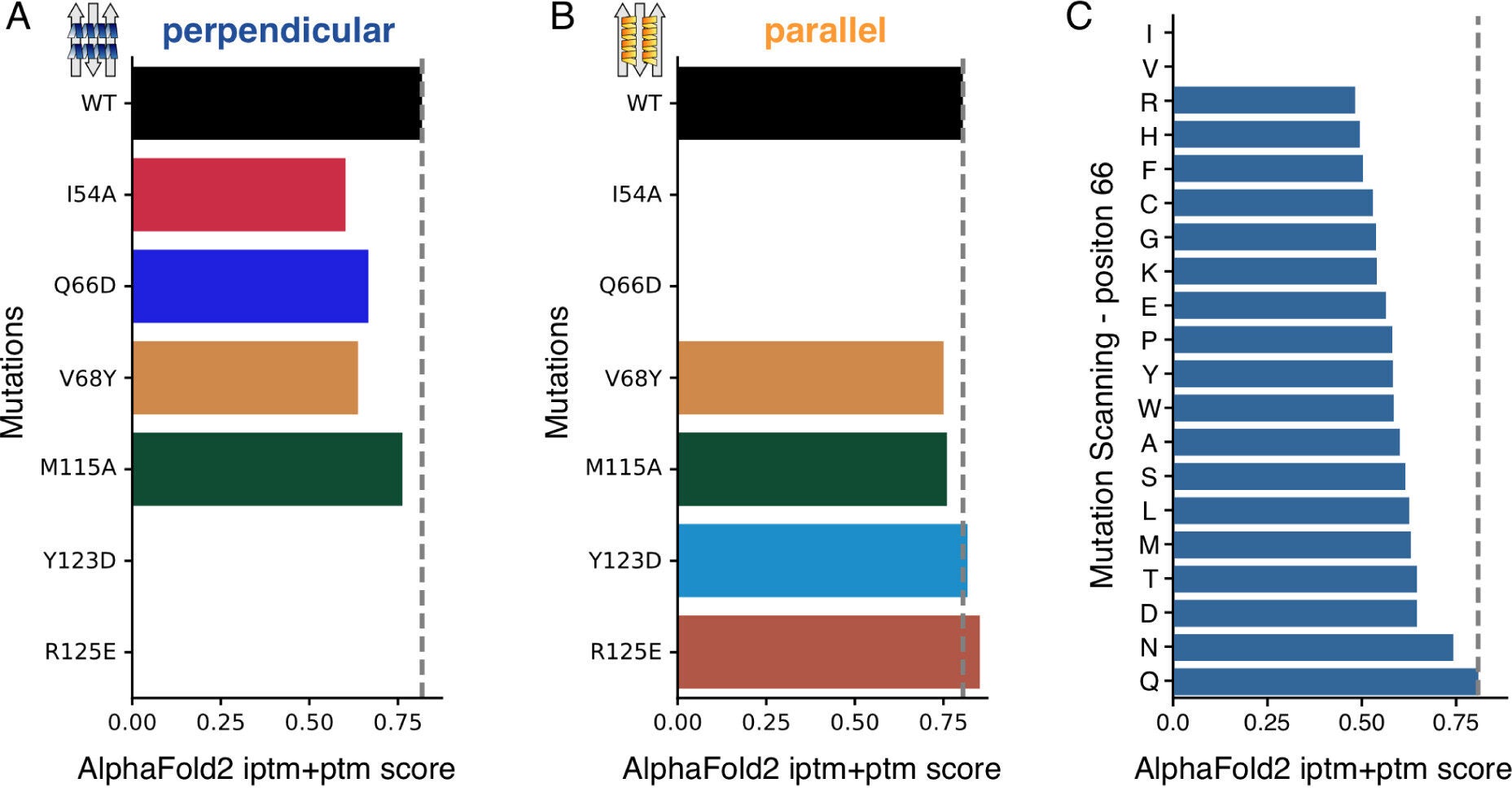
As the fight against cancer continues, integrating AI with dynamic network analysis and molecular dynamics will likely play a pivotal role in therapeutic innovation. Auburn University’s biophysics team exemplifies how collaboration across physics, chemistry, and biology can drive meaningful progress.
This research not only advances cancer treatment but also underscores the broader potential of computational tools in medicine. As Dr. Bernardi’s team continues to refine these techniques, the future of personalized and cost-effective therapies looks increasingly promising.
Note: Materials provided above by The Brighter Side of News. Content may be edited for style and length.
Like these kind of feel good stories? Get The Brighter Side of News’ newsletter.
The post Groundbreaking AI looks to revolutionize cancer treatments appeared first on The Brighter Side of News.