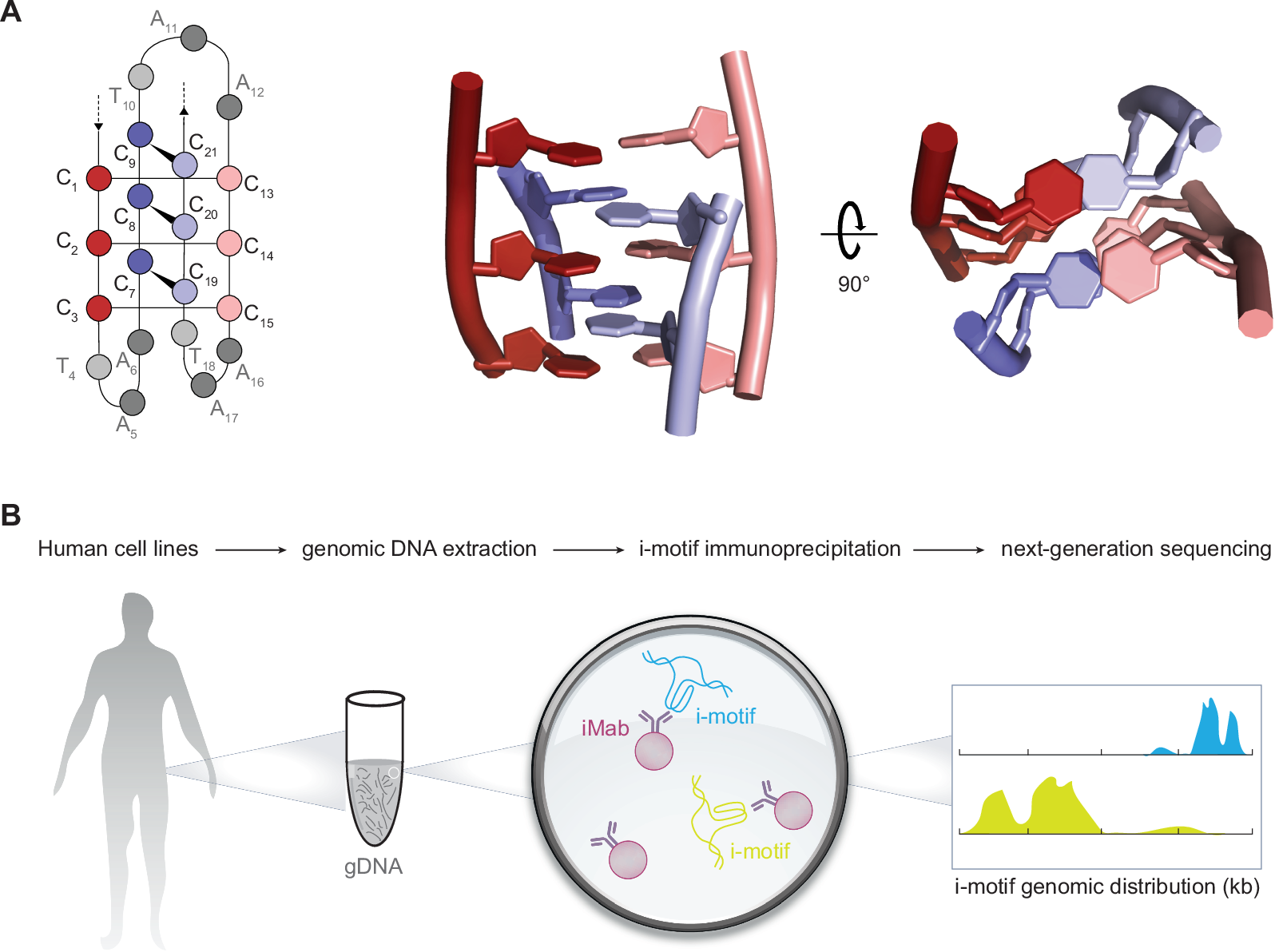
DNA’s iconic double helix has long been the symbol of life’s blueprint. However, the human genome conceals far more complexity. Among these mysteries are i-motifs, unusual knot-like DNA structures formed when cytosine-rich sequences fold into a four-stranded, twisted configuration.
Recent research has illuminated the prevalence and potential significance of i-motifs in human cells, shedding light on their possible role in gene regulation and disease.
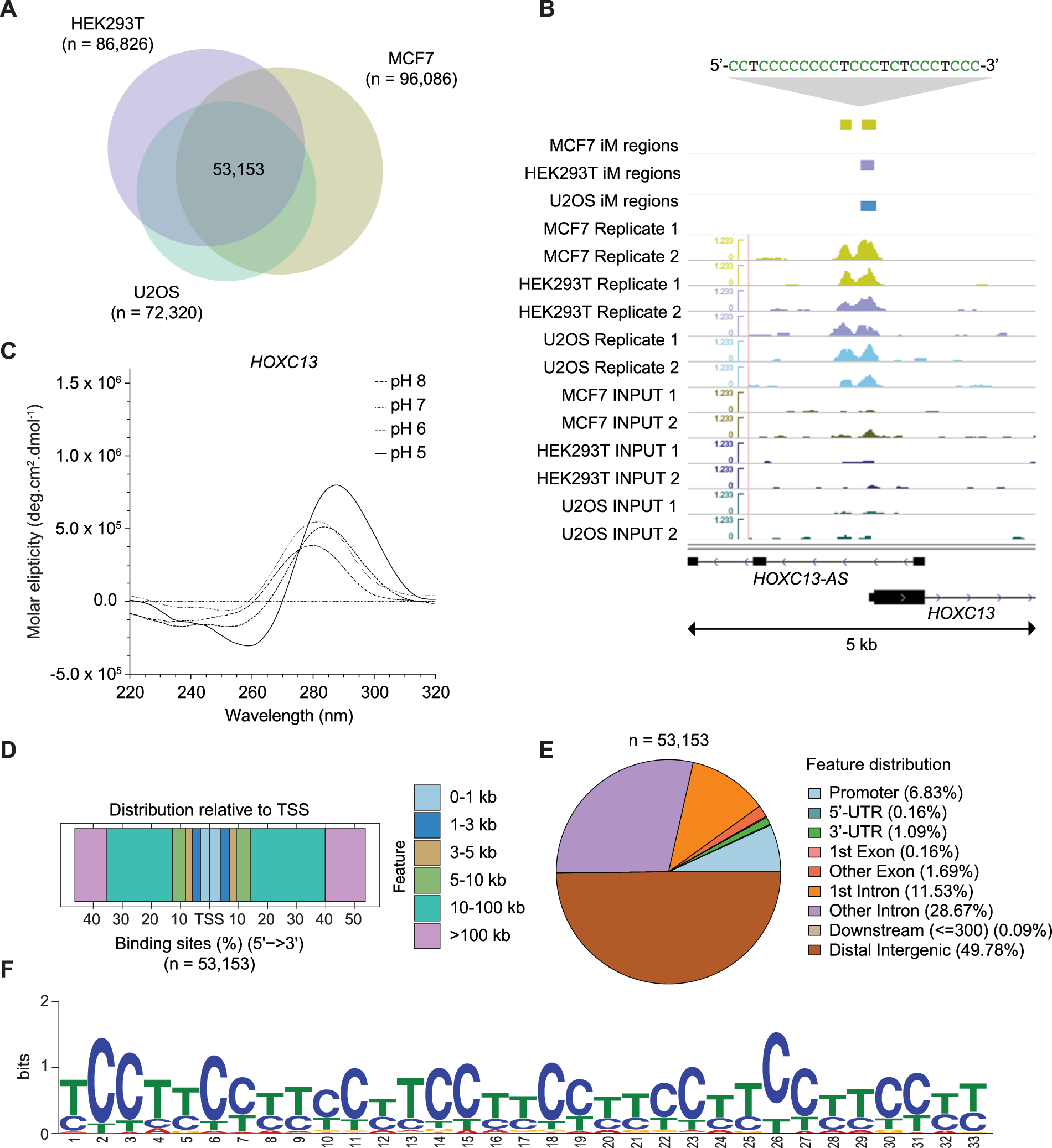
A groundbreaking study by researchers at the Garvan Institute of Medical Research has revealed the presence of more than 50,000 i-motifs across the human genome. This comprehensive mapping, published in The EMBO Journal, represents a major advance in understanding these mysterious structures.
Using a specially developed antibody capable of identifying and binding to i-motifs, researchers pinpointed their locations in three distinct human cell types.
Professor Daniel Christ, the study’s senior author, highlights the significance of these findings. “This study confirms that i-motifs are not just laboratory curiosities but widespread structures that likely play key roles in genomic function,” he says.
The discovery underscores how integral these formations may be to cellular processes, including gene regulation and cell cycle activity.
Unlike the canonical double helix, i-motifs form under specific conditions. They emerge when cytosine bases on the same DNA strand pair with each other, creating a unique structural feature.
Initially, scientists questioned their relevance due to their apparent dependence on acidic conditions for formation. However, subsequent research demonstrated that i-motifs could exist at physiological pH levels, especially under conditions like molecular crowding and DNA superhelicity.
Related Stories
The latest study found that i-motifs are not randomly distributed. Instead, they are concentrated in genomic regions critical for regulating gene activity. These areas include promoter regions of genes that are active during specific cell cycle phases.
“We discovered that i-motifs are associated with genes highly active during certain times in the cell cycle, suggesting they play a dynamic regulatory role,” explains Cristian David Peña Martinez, the study’s first author.
Intriguingly, i-motifs were also identified in the promoter regions of oncogenes, including the notoriously “undruggable” MYC oncogene. This discovery opens new avenues for targeting genes linked to cancer and other diseases.
The potential applications of i-motif research extend beyond understanding DNA structure. Their association with regulatory regions linked to disease highlights their promise as therapeutic and diagnostic targets.
Associate Professor Sarah Kummerfeld, a co-author of the study, underscores this potential. “The widespread presence of i-motifs near ‘holy grail’ sequences involved in hard-to-treat cancers opens up possibilities for innovative diagnostic and therapeutic approaches,” she says.
Designing drugs to target i-motifs could provide a novel way to influence gene expression, expanding options for treating diseases like cancer.
While the discovery of i-motifs marks a significant milestone, their full biological implications remain a subject of ongoing investigation. Advanced techniques, such as high-affinity i-motif immunoprecipitation followed by sequencing, have enabled researchers to map these structures and explore their genomic distribution. These tools have shown that i-motifs are prevalent in genes upregulated during the G0/G1 cell cycle phases.
“This study exemplifies how fundamental research and technological innovation can converge to make paradigm-shifting discoveries,” says Professor Christ. The ability to map i-motifs and identify their roles in gene activity provides a foundational resource for future studies.
Researchers aim to unravel their structural and molecular functions further, paving the way for breakthroughs in understanding genome architecture and function.
The journey to decode i-motifs continues, offering exciting opportunities to address fundamental questions about DNA’s hidden complexity.
As research progresses, these unique structures may hold the key to unlocking new treatments and diagnostics for diseases that have long eluded effective solutions.
Note: Materials provided above by The Brighter Side of News. Content may be edited for style and length.
Like these kind of feel good stories? Get The Brighter Side of News’ newsletter.
The post Researchers solve the mystery of 50,000 DNA “knots” in the human genome appeared first on The Brighter Side of News.